Compute likelihood profiles for a fitted model
# S3 method for glmmTMB
profile(
fitted,
parm = NULL,
level_max = 0.99,
npts = 8,
stepfac = 1/4,
stderr = NULL,
trace = FALSE,
parallel = c("no", "multicore", "snow"),
ncpus = getOption("profile.ncpus", 1L),
cl = NULL,
...
)
# S3 method for profile.glmmTMB
confint(object, parm = NULL, level = 0.95, ...)Arguments
- fitted
a fitted
glmmTMBobject- parm
which parameters to profile, specified
by index (position)
by name (matching the row/column names of
vcov(object,full=TRUE))as
"theta_"(random-effects variance-covariance parameters) or"beta_"(conditional and zero-inflation parameters)
- level_max
maximum confidence interval target for profile
- npts
target number of points in (each half of) the profile (approximate)
- stepfac
initial step factor (fraction of estimated standard deviation)
- stderr
standard errors to use as a scaling factor when picking step sizes to compute the profile; by default (if
stderrisNULL, orNAfor a particular element), uses the estimated (Wald) standard errors of the parameters- trace
print tracing information? If
trace=FALSEor 0, no tracing; iftrace=1, print names of parameters currently being profiled; iftrace>1, turn on tracing for the underlyingtmbprofilefunction- parallel
method (if any) for parallel computation
- ncpus
number of CPUs/cores to use for parallel computation
- cl
cluster to use for parallel computation
- ...
additional arguments passed to
tmbprofile- object
a fitted profile (
profile.glmmTMB) object- level
confidence level
Value
An object of class profile.glmmTMB, which is also a
data frame, with columns .par (parameter being profiled),
.focal (value of focal parameter), value (negative log-likelihood).
Details
Fits natural splines separately to the points from each half of the profile for each specified parameter (i.e., values above and below the MLE), then finds the inverse functions to estimate the endpoints of the confidence interval
Examples
if (FALSE) {
m1 <- glmmTMB(count~ mined + (1|site),
zi=~mined, family=poisson, data=Salamanders)
salamander_prof1 <- profile(m1, parallel="multicore",
ncpus=2, trace=1)
## testing
salamander_prof1 <- profile(m1, trace=1,parm=1)
salamander_prof1M <- profile(m1, trace=1,parm=1, npts = 4)
salamander_prof2 <- profile(m1, parm="theta_")
}
salamander_prof1 <- readRDS(system.file("example_files","salamander_prof1.rds",package="glmmTMB"))
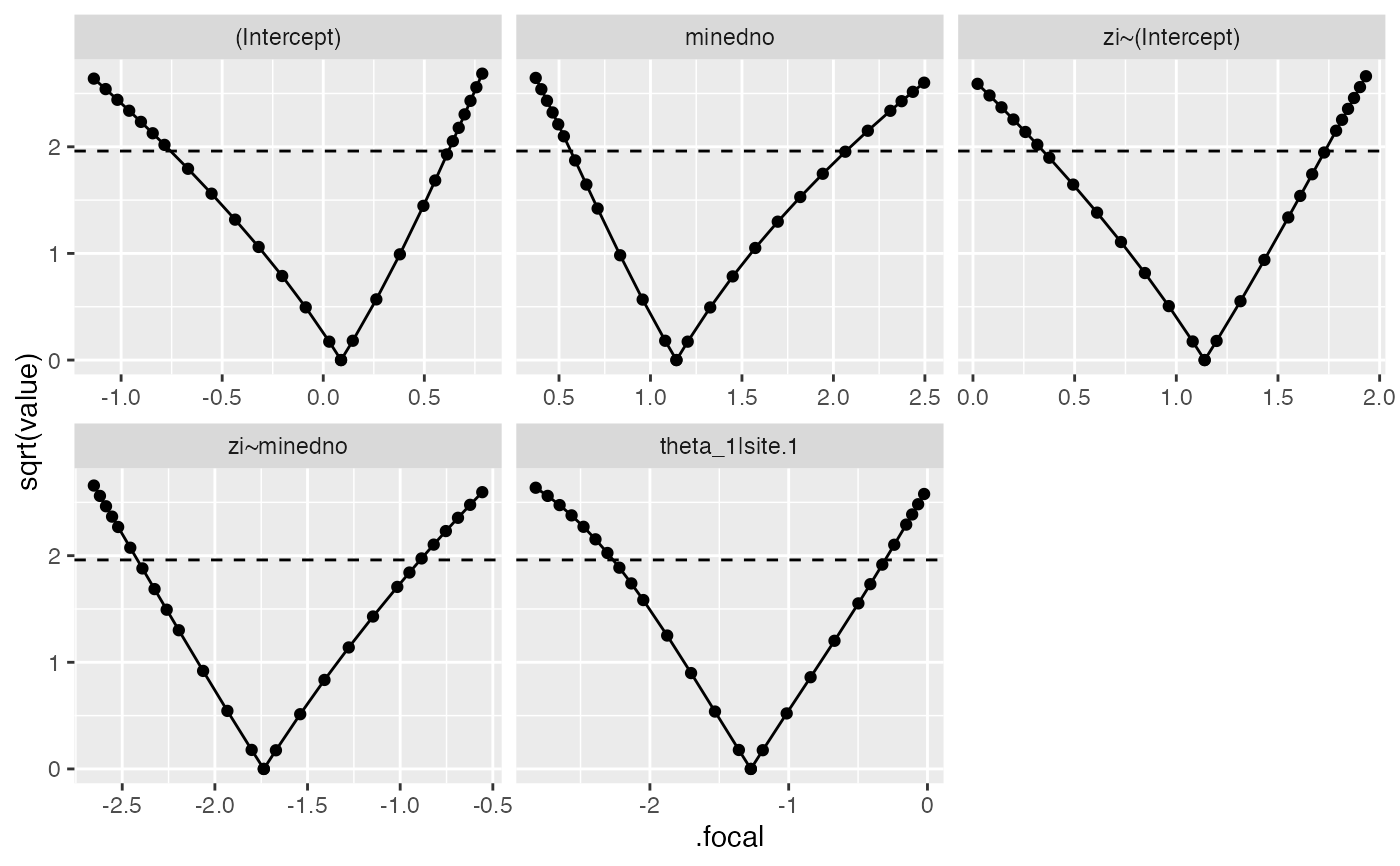
if (require("ggplot2")) {
ggplot(salamander_prof1,aes(.focal,sqrt(value))) +
geom_point() + geom_line()+
facet_wrap(~.par,scale="free_x")+
geom_hline(yintercept=1.96,linetype=2)
}
#> Loading required package: ggplot2
 salamander_prof1 <- readRDS(system.file("example_files","salamander_prof1.rds",package="glmmTMB"))
confint(salamander_prof1)
#> 2.5 % 97.5 %
#> (Intercept) -0.4686460 0.4805030
#> minedno 0.6449938 1.7417972
#> zi~(Intercept) 0.6088771 1.5648495
#> zi~minedno -2.2239195 -1.1665205
#> theta_1|site.1 -1.9452621 -0.5786442
confint(salamander_prof1,level=0.99)
#> 0.5 % 99.5 %
#> (Intercept) -0.6830922 0.5865649
#> minedno 0.6015779 1.9853898
#> zi~(Intercept) 0.4108581 1.6910201
#> zi~minedno -2.3713049 -0.9600516
#> theta_1|site.1 -2.1810585 -0.3697326
salamander_prof1 <- readRDS(system.file("example_files","salamander_prof1.rds",package="glmmTMB"))
confint(salamander_prof1)
#> 2.5 % 97.5 %
#> (Intercept) -0.4686460 0.4805030
#> minedno 0.6449938 1.7417972
#> zi~(Intercept) 0.6088771 1.5648495
#> zi~minedno -2.2239195 -1.1665205
#> theta_1|site.1 -1.9452621 -0.5786442
confint(salamander_prof1,level=0.99)
#> 0.5 % 99.5 %
#> (Intercept) -0.6830922 0.5865649
#> minedno 0.6015779 1.9853898
#> zi~(Intercept) 0.4108581 1.6910201
#> zi~minedno -2.3713049 -0.9600516
#> theta_1|site.1 -2.1810585 -0.3697326